https://doi.org/10.1021/acs.analchem.7b05157
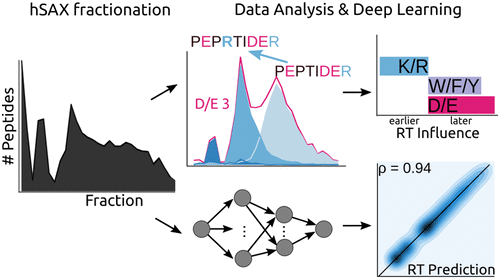
Hydrophilic strong anion exchange chromatography (hSAX) is becoming a popular method for the prefractionation of proteomic samples. However, the use and further development of this approach is affected by the limited understanding of its retention mechanism and the absence of elution time prediction. Using a set of 59 297 confidentially identified peptides, we performed an explorative analysis and built a predictive deep learning model. As expected, charged residues are the major contributors to the retention time through electrostatic interactions. Aspartic acid and glutamic acid have a strong retaining effect and lysine and arginine have a strong repulsion effect. In addition, we also find the involvement of aromatic amino acids. This suggests a substantial contribution of cation−π interactions to the retention mechanism. The deep learning approach was validated using 5-fold cross-validation (CV) yielding a mean prediction accuracy of 70% during CV and 68% on a hold-out validation set. The results of this study emphasize that not only electrostatic interactions but rather diverse types of interactions must be integrated to build a reliable hSAX retention time predictor.