doi: 10.1074/mcp.M115.048504
Abstract goes here
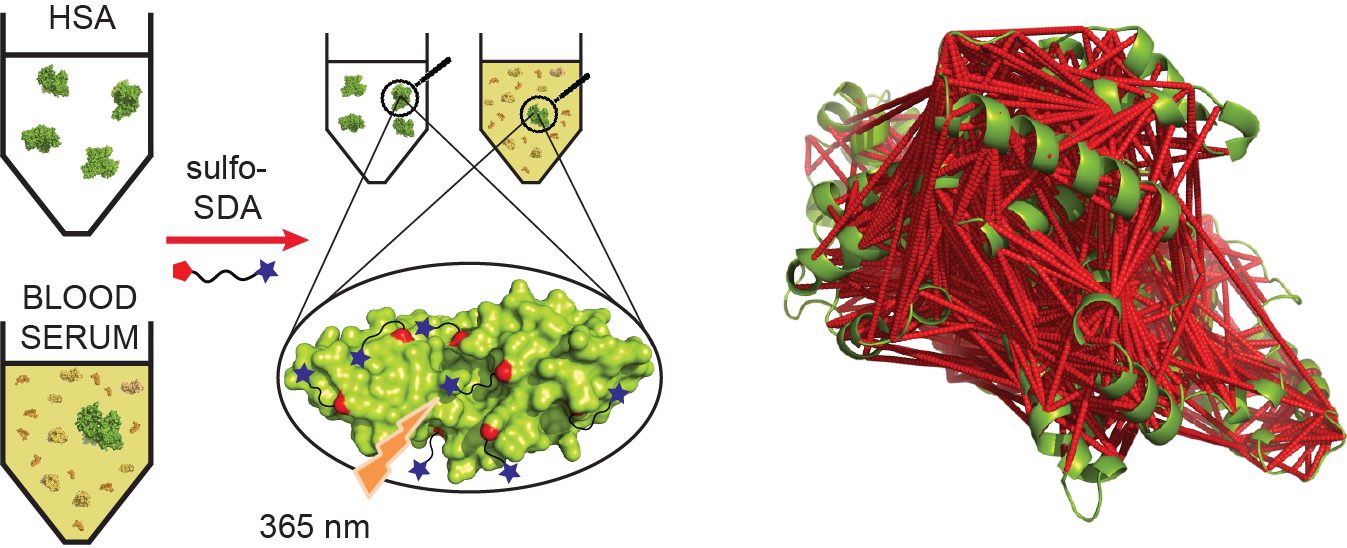
Chemical crosslinking combined with mass spectrometry has proven useful for studying protein-protein interactions and protein structure, however the low density of cross-link data has so far precluded its use in determining structures de novo. Crosslinking density has been typically limited by the chemical selectivity of the standard crosslinking reagents that are commonly used for protein crosslinking. We have implemented the use of a heterobifunctional crosslinking reagent, sulfosuccinimi- dyl 4,4-azipentanoate (sulfo-SDA), combining a traditional sulfo-N-hydroxysuccinimide (sulfo-NHS) ester and a UV photoactivatable diazirine group. This diazirine yields a highly reactive and promiscuous carbene species, the net result being a greatly increased number of crosslinks compared with homobifunctional, NHS-based crosslinkers. We present a novel methodology that combines the use of this high density photo-crosslinking data with conformational space search to investigate the structure of human serum albumin domains, from purified samples, and in its native environment, human blood serum. Our approach is able to determine human serum albumin domain structures with good accuracy: root-mean-square deviation to crystal structure are 2.8/5.6/2.9 Å (purified samples) and 4.5/5.9/4.8Å (serum samples) for domains A/B/C for the first selected structure; 2.5/4.9/2.9 Å (purified samples) and 3.5/5.2/3.8 Å (serum samples) for the best out of top five selected structures. Our proof-of-concept study on human serum albumin demonstrates initial potential of our approach for determining the structures of more proteins in the complex biological contexts in which they function and which they may require for correct folding. Data are available via ProteomeXchange with identifier PXD001692.