Lars Kolbowski, Adam Belsom, Juri Rappsilber
Journal of the American Society for Mass Spectrometry, 2020
https://doi.org/10.1021/jasms.0c00106
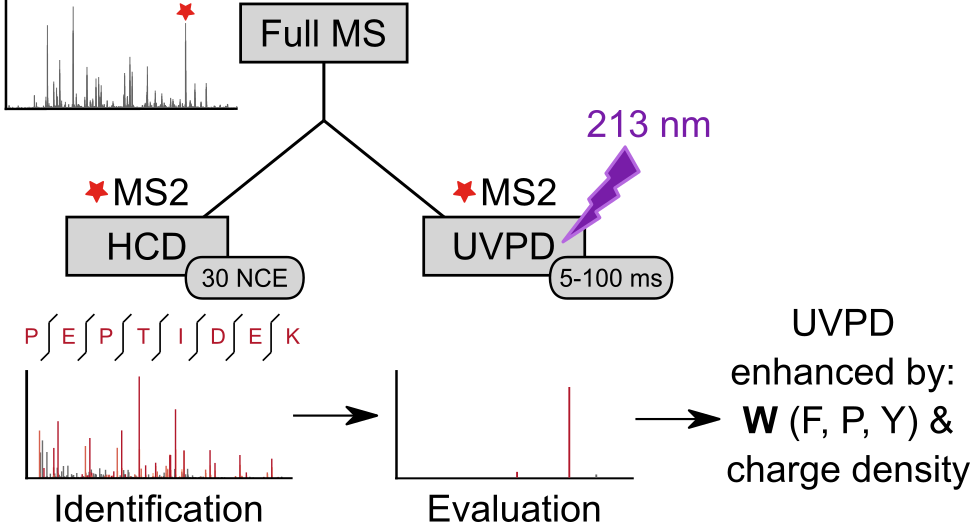
We analyzed the backbone fragmentation behavior of tryptic peptides of a four protein mixture and of E. coli lysate subjected to Ultraviolet Photodissociation (UVPD) at 213 nm on a commercially available UVPD-equipped tribrid mass spectrometer. We obtained 15,178 unique high-confidence peptide UVPD spectrum matches by re-cording a reference beam-type collision-induced dissociation (HCD) spectrum of each precursor, ensuring our investigation includes a broad selection of peptides, including those that fragmented poorly by UVPD. Type a, b and y ions were most prominent in UVPD spectra and median sequence coverage ranged from 5.8% (at 5 ms laser excitation time) to 45.0% (at 100 ms). Overall sequence fragment intensity remained relatively low (median: 0.4% (5 ms) to 16.8% (100 ms) of total intensity) and remaining precursor intensity high. Sequence coverage and sequence fragment intensity ratio correlated with precursor charge density, suggesting that UVPD at 213 nm may suffer from newly formed fragments sticking together due to non-covalent interactions. UVPD fragmentation efficiency therefore might benefit from supplemental activation, as was shown for ETD. Aromatic amino acids, most prominently tryptophan, facilitated UVPD. This points at aromatic tags as possible enhancers of UVPD. Data are available via ProteomeXchange with identifier PXD018176 and on spectrumview-er.org/db/UVPD-213nm-trypPep.